Nexus User Scripts
Users interact with Nexus by writing a Python script that often resembles an input file. A Nexus user script typically consists of six main sections, as described below:
- Nexus imports:
functions unique to Nexus are drawn into the user environment.
- Nexus settings:
specify machine information and configure the runtime behavior of Nexus.
- Physical system specification:
create a data description of the physical system. Generate a crystal structure or import one from an external data file. Physical system details can be shared among many simulations.
- Workflow specification:
describe the simulations to be performed. Link simulations together by their data dependencies to form workflows.
- Workflow execution:
pass control to Nexus for active workflow management. Simulation input files are generated, jobs are submitted and monitored, output files are collected and preprocessed for later analysis.
- Data analysis:
control returns to the user to extract preprocessed simulation output data for further analysis.
Each of the six input sections is the subject of lengthier discussion: Nexus imports, Nexus settings: global state and user-specific information, Physical system specification, Workflow specification, Workflow execution, and Data analysis. These sections are also illustrated in the abbreviated example script below. For more complete examples and further discussion, please refer to the user walkthroughs in Complete Examples.
Nexus imports
Each script begins with imports from the main Nexus module. Items imported include the interface to provide settings to Nexus, helper functions to make objects representing atomic structures or simulations of particular types (e.g. QMCPACK or VASP), and the interface to provide simulation workflows to Nexus for active management.
The import of all Nexus components is accomplished with the brief
“from nexus import *”. Each component can also be imported
separately by name, as in the example below.
from nexus import settings # Nexus settings function
from nexus import generate_physical_system # for creating atomic structures
from nexus import generate_pwscf # for creating PWSCF sim. objects
from nexus import Job # for creating job objects
from nexus import run_project # for active workflow management
This has the advantage of avoided unwanted namespace collisions with user defined variables. The major Nexus components available for import are listed in Table 1.
component |
description |
|---|---|
|
Alter runtime behavior. Provide machine information. |
|
Create atomic structure including electronic information. |
|
Create atomic structure without electronic information. |
|
Create generic simulation object. |
|
Create PWSCF simulation object. |
|
Create VASP simulation object. |
|
Create GAMESS simulation object. |
|
Create QMCPACK simulation object. |
|
Create SQD simulation object. |
|
Create generic input file object. |
|
Create generic input file object representing multiple files. |
|
Create PWSCF input file object. |
|
Create VASP input file object. |
|
Create GAMESS input file object. |
|
Create QMCPACK input file object. |
|
Create SQD input file object. |
|
Provide job information for simulation run. |
|
Initiate active workflow management. |
|
Generic container object. Store inputs for later use. |
Nexus settings: global state and user-specific information
Following imports, the next section of a Nexus script is dedicated to
providing information regarding the local machine, the location of
various files, and the desired runtime behavior. This information is
communicated to Nexus through the settings function. To make
settings available in your project script, use the following import
statement:
from nexus import settings
In most cases, it is sufficient to supply only four pieces of
information through the settings function: whether to run all jobs
or just create the input files, how often to check jobs for completion,
the location of pseudopotential files, and a description of the local
machine.
settings(
generate_only = True, # only write input files, do not run
sleep = 3, # check on jobs every 3 seconds
pseudo_dir = './pseudopotentials', # path to PP file collection
machine = 'ws8' # local machine is an 8 core workstation
)
A few additional parameters are available in settings to control
where runs are performed, where output data is gathered, and whether to
print job status information. More detailed information about machines
can be provided, such as allocation account numbers, filesystem
structure, and where executables are located.
settings(
status_only = True, # only show job status, do not write or run
generate_only = True, # only write input files, do not run
sleep = 3, # check on jobs every 3 seconds
pseudo_dir = './pseudopotentials', # path to PP file collection
runs = '', # base path for runs is local directory
results = '/home/jtk/results/', # light output data copied elsewhere
machine = 'titan', # Titan supercomputer
account = 'ABC123', # user account number
)
Physical system specification
After providing settings information, the user often defines the atomic structure to be studied (whether generated or read in). The same structure can be used to form input to various simulations (e.g. DFT and QMC) performed on the same system. The examples below illustrate the main options for structure input.
Read structure from a file
dia16 = generate_physical_system(
structure = './dia16.POSCAR', # load a POSCAR file
C = 4 # pseudo-carbon (4 electrons)
)
Generate structure directly
dia16 = generate_physical_system(
lattice = 'cubic', # cubic lattice
cell = 'primitive', # primitive cell
centering = 'F', # face-centered
constants = 3.57, # a = 3.57
units = 'A', # Angstrom units
atoms = 'C', # monoatomic C crystal
basis = [[0,0,0], # basis vectors
[.25,.25,.25]], # in lattice units
tiling = (2,2,2), # tile from 2 to 16 atom cell
C = 4 # pseudo-carbon (4 electrons)
)
Provide cell, elements, and positions explicitly:
dia16 = generate_physical_system(
units = 'A', # Angstrom units
axes = [[1.785,1.785,0. ], # cell axes
[0. ,1.785,1.785],
[1.785,0. ,1.785]],
elem = ['C','C'], # atom labels
pos = [[0. ,0. ,0. ], # atomic positions
[0.8925,0.8925,0.8925]],
tiling = (2,2,2), # tile from 2 to 16 atom cell
kgrid = (4,4,4), # 4 by 4 by 4 k-point grid
kshift = (0,0,0), # centered at gamma
C = 4 # pseudo-carbon (4 electrons)
)
In each of these cases, the text “C = 4” refers to the number of
electrons in the valence for a particular element. Here a
pseudopotential is being used for carbon and so it effectively has four
valence electrons. One line like this should be included for each
element in the structure.
Workflow specification
The next section in a Nexus user script is the specification of simulation workflows. This stage can be logically decomposed into two sub-stages: (1) specifying inputs to each simulation individually, and (2) specifying the data dependencies between simulations.
Generating simulation objects
Simulation objects are created through calls to “generate_xxxxxx”
functions, where “xxxxxx” represents the name of a particular
simulation code, such as pwscf, vasp, or qmcpack. Each
generate function shares certain inputs, such as the path where the
simulation will be performed, computational resources required by the
simulation job, an identifier to differentiate between simulations (must
be unique only for simulations occurring in the same directory), and the
atomic/electronic structure to simulate:
relax = generate_pwscf(
identifier = 'relax', # identifier for the run
path = 'diamond/relax', # perform run at this location
job = Job(cores=16,app='pw.x'), # run on 16 cores using pw.x executable
system = dia16, # 16 atom diamond cell made earlier
pseudos = ['C.BFD.upf'], # pseudopotential file
files = [], # any other files to be copied in
... # PWSCF-specific inputs follow
)
The simulation objects created in this way are just data. They represent requests for particular simulations to be carried out at a later time. No simulation runs are actually performed during the creation of these objects. A basic example of generation input for each of the four major codes currently supported by Nexus is given below.
Quantum Espresso (PWSCF) generation:
scf = generate_pwscf(
identifier = 'scf',
path = 'diamond/scf',
job = scf_job,
system = dia16,
pseudos = ['C.BFD.upf'],
input_type = 'generic',
calculation = 'scf',
input_dft = 'lda',
ecutwfc = 75,
conv_thr = 1e-7,
kgrid = (2,2,2),
kshift = (0,0,0),
)
The keywords calculation, input_dft, ecutwfc, and
conv_thr will be familiar to the casual user of PWSCF. Any input
keyword that normally appears as part of a namelist in PWSCF input can
be directly supplied here. The generate_pwscf function, like most of
the others, actually takes an arbitrary number of keyword arguments.
These are later screened against known inputs to PWSCF to avoid errors.
The kgrid and kshift inputs inform the KPOINTS card in the
PWSCF input file, overriding any similar information provided in
generate_physical_system.
VASP generation:
relax = generate_vasp(
identifier = 'relax',
path = 'diamond/relax',
job = relax_job,
system = dia16,
pseudos = ['C.POTCAR'],
input_type = 'generic',
istart = 0,
icharg = 2,
encut = 450,
nsw = 5,
ibrion = 2,
isif = 2,
kcenter = 'monkhorst',
kgrid = (2,2,2),
kshift = (0,0,0),
)
Similar to generate_pwscf, generate_vasp accepts an arbitrary
number of keyword arguments and any VASP input file keyword is accepted
(the VASP keywords provided here are istart, icharg, encut,
nsw, ibrion, and isif). The kcenter, kgrid, and
kshift keywords are used to form the KPOINTS input file.
Pseudopotentials provided through the pseudos keyword will fused
into a single POTCAR file following the order of the atoms created
by generate_physical_system.
GAMESS generation:
uhf = generate_gamess(
identifier = 'uhf',
path = 'water/uhf',
job = Job(cores=16,app='gamess.x'),
system = h2o,
pseudos = [H.BFD.gms,O.BFD.gms],
symmetry = 'Cnv 2',
scftyp = 'uhf',
runtyp = 'energy',
ispher = 1,
exetyp = 'run',
maxit = 200,
memory = 150000000,
guess = 'hcore',
)
The generate_gamess function also accepts arbitrary GAMESS keywords
(symmetry, scftyp, runtyp, ispher, exetyp,
maxit, memory, and guess here). The pseudopotential files
H.BFD.gms and O.BFD.gms include the gaussian basis sets as well
as the pseudopotential channels (the two parts are just concatenated
into the same file, commented lines are properly ignored). Nexus drives
the GAMESS executable (gamess.x here) directly without the
intermediate rungms script as is often done. To do this, the
ericfmt keyword must be provided in settings specifying the path
to ericfmt.dat.
QMCPACK generation:
qmc = generate_qmcpack(
identifier = 'vmc',
path = 'diamond/vmc',
job = Job(cores=16,threads=4,app='qmcpack'),
input_type = 'basic',
system = dia16,
pseudos = ['C.BFD.xml'],
jastrows = [],
calculations = [
vmc(
walkers = 1,
warmupsteps = 20,
blocks = 200,
steps = 10,
substeps = 2,
timestep = .4
)
],
dependencies = (conv,'orbitals')
)
Unlike the other generate functions, generate_qmcpack takes only
selected inputs. The reason for this is that QMCPACK’s input file is
highly structured (nested XML) and cannot be directly mapped to
keyword-value pairs. The full set of allowed keywords is beyond the
scope of this section. Please refer to the user walkthroughs provided in
Complete Examples for further examples.
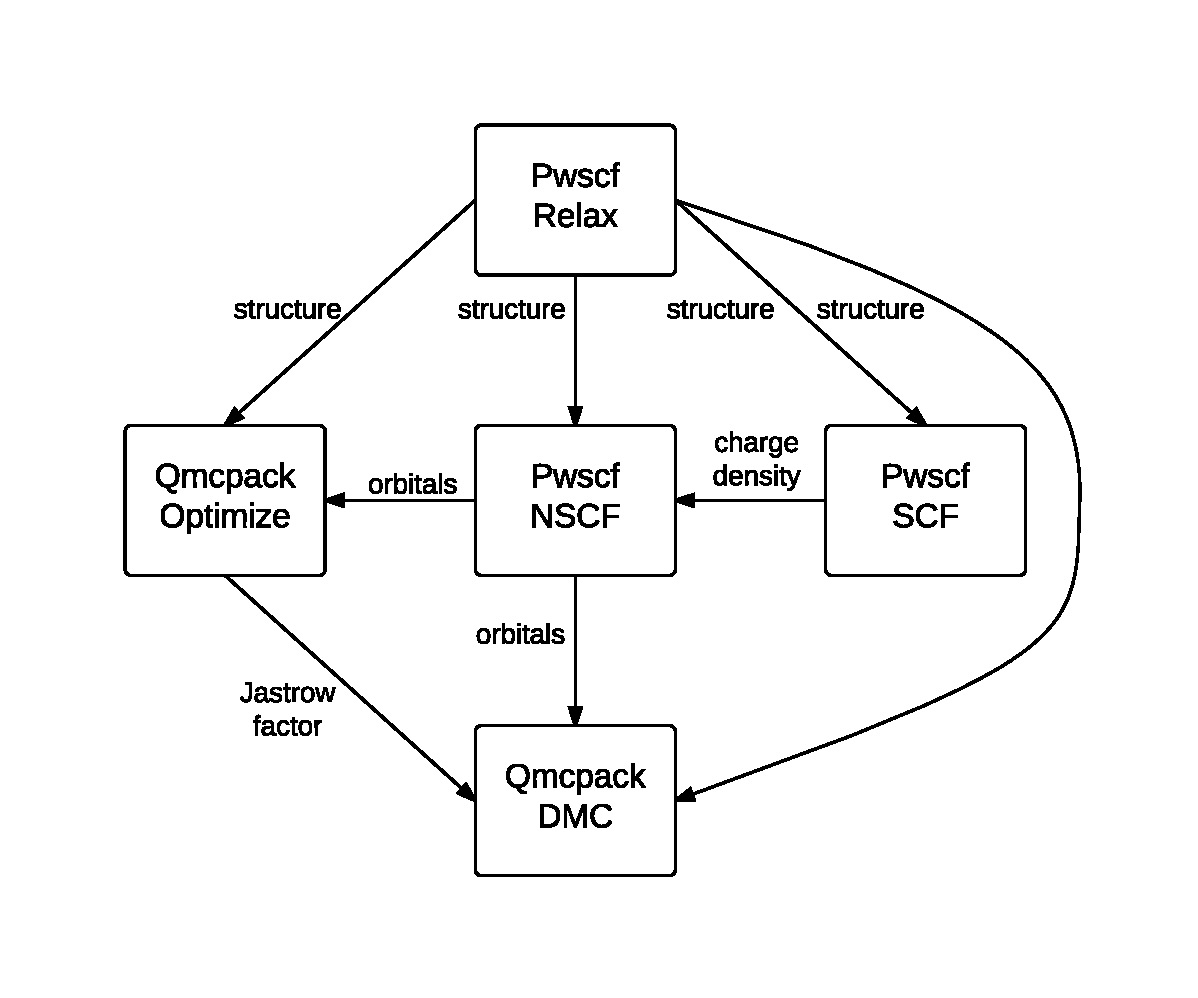
Fig. 1 An example Nexus workflow/cascade involving QMCPACK and PWSCF. The arrows and labels denote the flow of information between the simulation runs.
Composing workflows from simulation objects
Simulation workflows are created by specifying the data dependencies between simulation runs. An example workflow is shown in Fig. 1. In this case, a single relaxation calculation performed with PWSCF is providing a relaxed structure to each of the subsequent simulations. PWSCF is used to create a converged charge density (SCF) and then orbitals at specific k-points (NSCF). These orbitals are used by each of the two QMCPACK runs; the first optimization run provides a Jastrow factor to the final DMC run.
Below is an example of how this workflow can be created with Nexus. Most
keywords to the generate functions have been omitted for brevity.
The conv step listed below is implicit in Fig. 1.
relax = generate_pwscf(
...
)
scf = generate_pwscf(
dependencies = (relax,'structure'),
...
)
nscf = generate_pwscf(
dependencies = [(relax,'structure' ),
(scf ,'charge_density')],
...
)
conv = generate_pw2qmcpack(
dependencies = (nscf ,'orbitals' ),
...
)
opt = generate_qmcpack(
dependencies = [(relax,'structure'),
(conv ,'orbitals' )],
...
)
dmc = generate_qmcpack(
dependencies = [(relax,'structure'),
(conv ,'orbitals' ),
(opt ,'jastrow' )],
...
)
As suggested at the beginning of this section, workflow composition logically breaks into two parts: simulation generation and workflow dependency specification. This type of breakup can also be performed explicitly within a Nexus user script, if desired:
# simulation generation
relax = generate_pwscf(...)
scf = generate_pwscf(...)
nscf = generate_pwscf(...)
conv = generate_pw2qmcpack(...)
opt = generate_qmcpack(...)
dmc = generate_qmcpack(...)
# workflow dependency specification
scf.depends(relax,'structure')
nscf.depends((relax,'structure' ),
(scf ,'charge_density'))
conv.depends(nscf ,'orbitals' )
opt.depends((relax,'structure'),
(conv ,'orbitals' ))
dmc.depends((relax,'structure'),
(conv ,'orbitals' ),
(opt ,'jastrow' ))
More complicated workflows or scans over parameters of interest can be created with for loops and if-else logic constructs. This is fairly straightforward to accomplish because any keyword input can given a Python variable instead of a constant, as is mostly the case in the brief examples above.
Workflow execution
Simulation jobs are actually executed when the corresponding simulation
objects are passed to the run_project function. Within the
run_project function, most of the workflow management operations
unique to Nexus are actually performed. The details of the management
process is not the purpose of this section. This process is discussed in
context in the Complete Examples walkthroughs.
The run_project function can be invoked in a couple of ways. The
most straightforward is simply to provide all simulation objects
directly as arguments to this function:
run_project(relax,scf,nscf,opt,dmc)
When complex workflows are being created (e.g. when the generate
function appear in for loops and if statements), it is generally
more convenient to accumulate a list of simulation objects and then pass
the list to run_project as follows:
sims = []
relax = generate_pwscf(...)
sims.append(relax)
scf = generate_pwscf(...)
sims.append(scf)
nscf = generate_pwscf(...)
sims.append(nscf)
conv = generate_pw2qmcpack(...)
sims.append(conv)
opt = generate_qmcpack(...)
sims.append(opt)
dmc = generate_qmcpack(...)
sims.append(dmc)
run_project(sims)
When the run_project function returns, all simulation runs should be
finished.
Limiting the number of submitted jobs
Nexus will submit all eligible jobs at the same time unless told otherwise. This can be a large number when many calculations are present within the same project, e.g. various geometries or twists. While this is fine on local resources, it might break the rules at computing centers such as ALCF where only 20 jobs can be submitted at the same time. In such cases, it is possible to specify the the size of the queue in Nexus to avoid monopolizing the resources.
from nexus import get_machine
theta = get_machine('theta')
theta.queue_size = 10
In this case, Nexus will never submit more than 10 jobs at a time, even if more jobs are ready to be submitted, or resources on the local machine are available.
Having the option of limiting the number of jobs running at the same time can be useful even on local workstations (to avoid taking over all the available resources). In such a case, a simpler strategy is possible by claiming fewer available cores in settings, e.g. machine=’ws8’ vs ‘ws4’ vs ‘ws2’ etc.
Data analysis
Following the call to run_project, the user can perform data
analysis tasks, if desired, as the analyzer object associated with each
simulation contains a collection of post-processed output data rendered
in numeric form (ints, floats, numpy arrays) and stored in a structured
format. An interactive example for QMCPACK data analysis is shown below.
Note that all analysis objects are interactively browsable in a similar
manner.
>>> qa=dmc.load_analyzer_image()
>>> qa.qmc
0 VmcAnalyzer
1 DmcAnalyzer
2 DmcAnalyzer
>>> qa.qmc[2]
dmc DmcDatAnalyzer
info QAinformation
scalars ScalarsDatAnalyzer
scalars_hdf ScalarsHDFAnalyzer
>>> qa.qmc[2].scalars_hdf
Coulomb obj
ElecElec obj
Kinetic obj
LocalEnergy obj
LocalEnergy_sq obj
LocalPotential obj
data QAHDFdata
>>> print qa.qmc[2].scalars_hdf.LocalEnergy
error = 0.0201256357883
kappa = 12.5422841447
mean = -75.0484800012
sample_variance = 0.00645881103012
variance = 0.850521272106